The Genomes To Fields (G2F) Initiative
A publicly initiated and led research initiative to catalyze and coordinate research linking genomics and predictive phenomics to achieve advances that generate societal and environmental benefits
Background
G2F is an umbrella initiative to support translation of maize genomic information for the benefit of growers, consumers and society. This public-private partnership is building on publicly funded corn genome sequencing projects to develop approaches to understand the functions of corn genes and specific alleles across environments. Ultimately this information will be used to enable to accurate prediction of the phenotypes of corn plants in diverse environments. There are many dimensions to the over-arching goal of understanding genotype-by-environment (GxE) interactions, including which genes impact which traits and trait components, how genes interact among themselves (GxG), the relevance of specific genes under different growing conditions, and how these genes influence plant growth during various stages of development. This Initiative will promote projects that advance integrated research and technologies, combining fields such as genetics, genomics, plant physiology, agronomy, climatology and crop modeling, with computation and informatics, statistics and engineering. Scientists from these non-agricultural fields are being invited to participate in G2F.
The initial concept for Genomes to Fields was developed in 2013. Since then the Iowa Corn Promotion Board has invested resources to jump-start this Initiative. Financial support will be sought from public funding agencies. The seed industry is also being invited to participate in and benefit from the Initiative. In addition, other types of companies that could benefit from this Initiative are being invited to participate.
The initiative is developing a strategy to work closely with Congress and government agencies to obtain research funding.
Several projects are already underway:
G2F Executive Committee
Project: Genomes by Environment (GxE)
Since 2014, this project has evaluated approximately 180,000 field plots involving more than 2500 corn hybrid varieties across 162 unique environments in North America.
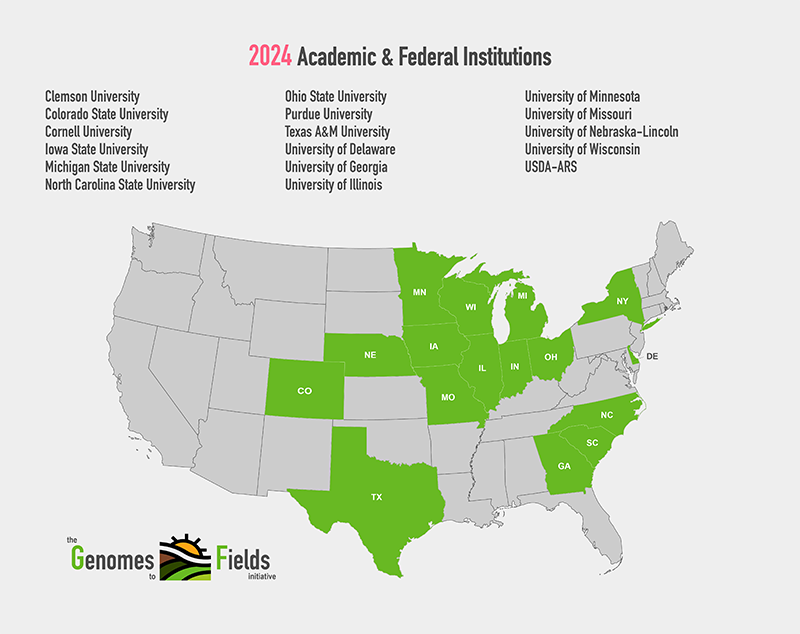
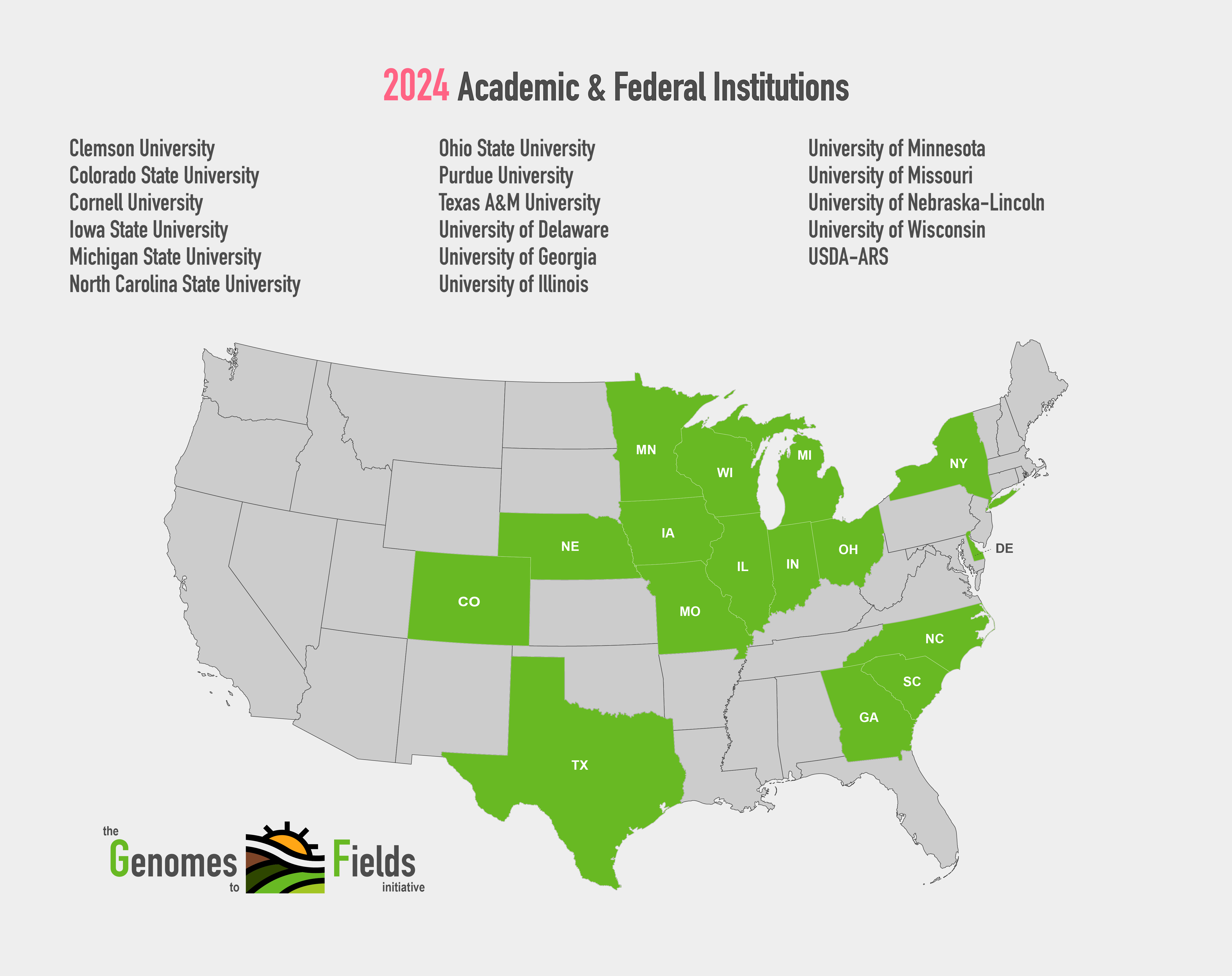
2024 GxE Map showing academic and federal institutions
View Enlarged MapCore Traits
Plant
morphology
Plant Height
Ear Height
Ear
morphology
Ear width
Ear length
Cob width
Kernel row number
Agronomic
Stand count
Root lodging
Stalk lodging
Days to silking/anthesis
Productivity
Grain moisture
Test weight
Grain yield
Collaborators and sponsors: (sorted by last name in ascending order)
Tim Beissinger (Göttingen)
Martin Bohn (UIUC)
Ed Buckler (ARS)
Darwin Campbell (ISU)
Alejandro Castro (UW)
Ignacio Ciampitti (KSU)
Liang Dong (ISU)
Jode Edwards (ARS)
David Ertl (IA Corn)
Sherry Flint-Garcia (ARS)
Joseph Gage (Cornell)
Jack Gardiner (ISU)
Fiona Goggin (AR State)
Byron Good (Guelph)
Mike Gore (Cornell)
Christopher Graham (SDSU)
Patricio Grassini (UNL)
Jerry Hatfield (ARS)
Candy Hirsch (UMN)
Jim Holland (ARS)
Elizabeth Hood (AR State)
David Hooker (Guelph)
Diego Jarquin (UNL)
Shawn Kaeppler (UW)
Aida Kebede (AAFC)
Joe Knoll (ARS)
Greg Kruger (UNL)
Nick Lauter (ARS)
Carolyn Lawrence-Dill (ISU)
Liz Lee (Guelph)
Dayane Cristina Lima (UW)
Sanzchen Liu (Kansas)
Argelia Lorence (AR State)
Aaron Lorenz (UMN)
Jonathan Lynch (PSU)
Bridget McFarland (UW)
John McKay (Colorado)
Nathan Miller (UW)
Richard Minyo (OSU)
Steve Moose (UIUC)
Seth Murray (TAMU)
Rebecca Nelson (Cornell)
Osler Ortez (OSU)
Torbert Rocheford (Purdue)
Oscar Rodriguez (UNL)
Cinta Romay (Cornell)
James Schnable (UNL)
Pat Schnable (ISU)
Brian Scully (ARS)
Rajandeep Sekhon (Clemson)
Kevin Silverstein (UMN)
Maninder Singh (MI State)
Margaret Smith (Cornell)
Edgar Spalding (UW)
Erin Sparks (Univ. Delaware)
Nathan Springer (UMN)
Yiwei Sun (ISU)
Kurt Thelen (MSU)
Peter Thomison (OSU)
Addie Thompson (MI State)
Kelly Thorp (ARS)
Mitch Tuinstra (Purdue)
Jason Wallace (UGA)
Jacob Washburn (USDA)
Rod Williamson (IA Corn)
Randy Wisser (UDel)
Wenwei Xu (TAMU)
Jianming Yu (ISU)
Natalia de Leon (UW)
The G2F network comprises public sector collaborators in the US and Canada. G2F often shares public germplasm and our best practices with public-sector researchers outside of the US and Canada as well as those in the private sector. If you'd like to discuss possibilities please contact G2F Executive Committee.
Project: Information Management Resources
Data Curation and Next-Gen Analytics: Working closely with colleagues at the University of Wisconsin, Madison and the University of Minnesota team from G.E.M.S™, an innovative agroinformatics, data discovery, sharing and analytics platform, is planning, coordinating, and implementing data management for the GxE subgroup. Working together, we plan to streamline data collection and submission, enable real-time data cleaning upon upload with real-time analysis and comparison to previous-years' data. Activities are expected to grow to incorporate transformative technologies in routine analyses, such as real-time incorporation of weather data, UAV, satellite, and other remote-sensed field data, and in-season predictive analytics.
Development of Core Trait Descriptions: In collaboration with Martin Bohn, a list of 14 Core Traits with the highest priorities for measurement was assembled and distributed to the GxE cooperators. Specific details on the measurement of each trait included trait abbreviations, units of measurement, measurement timing, accuracy requirements, and metadata considerations. Core Trait data were documented using standards that enable complex observations to be calculated. For example, dates of planting as well as dates of silking and anthesis were recorded rather than “Days to silking/anthesis”. This also enables integration with weather and other environmental data.
Two Phenotypic Data Types – Breeding Measurements and High-throughput Images: Methods for breeding measurements have long been described. Standards being deployed for the GxE subgroup are based on expanding the CIMMYT trait descriptors. For emerging data types like high-throughput images, the field is in its infancy and it is anticipated that deploying specific and required standards would impede the development of novel and revolutionary data documentation techniques. For this reason, multiple standards and platforms for high-throughput image data are envisioned, planned, and supported.
Weather Data Collection: Spectrum weather stations are present at all GxE locations. Detailed weather data (10 measurements) was collected every 30 minutes for the entire season through harvest and deposited along with trait data and metadata for each GxE cooperator.